vignettes/Tutorials/Model_simulation.Rmd
Model_simulation.RmdHow to simulate in DDModeling
To simulate data in DDModeling, you need a specified DDModel (see here for a tutorial).
library(DDModeling) #> Lade nötiges Paket: data.table DSTP <- DDModel(model="DSTP",task = "flanker",CDF_perc = c(0.1,0.3,0.5,0.7,0.9),CAF_perc = c(0.0,0.2,0.4,0.6,0.8,1.0))
With this model the simulation using Sim_DDModel() is uncomplicated.
some_data <- Sim_DDModel(model=DSTP,trials = 1000L)
Sim_DDModel() requires at least two arguments model and trials. The former specifies a DDModel object, the latter the number of trials per condition to be simulated. It then returns a DDRep object, which can be inspected by simply calling it:
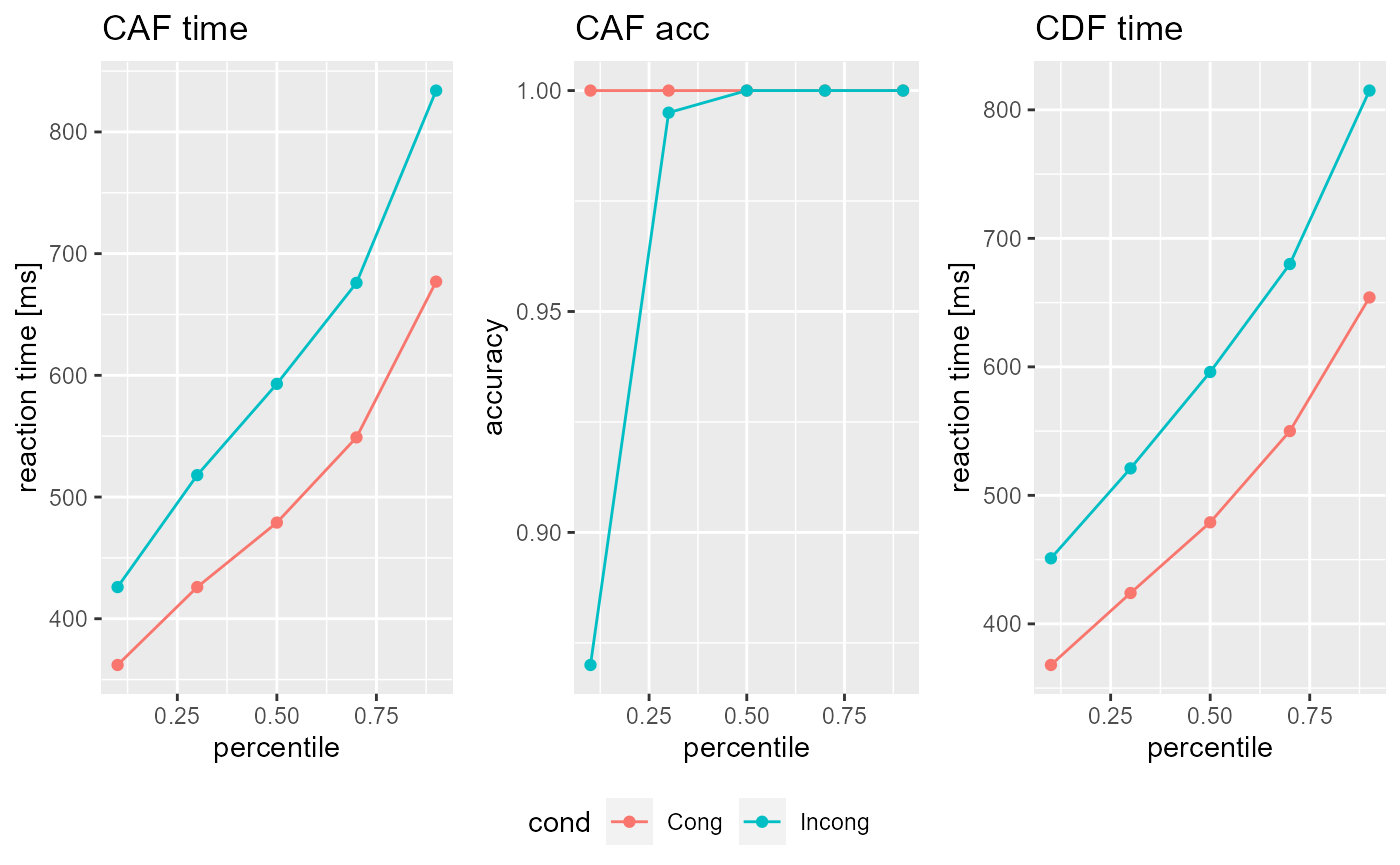
some_data #> CDF: #> $Cong #> cond perc time N #> 1 Cong 0.1 368 100 #> 2 Cong 0.3 424 300 #> 3 Cong 0.5 479 500 #> 4 Cong 0.7 550 700 #> 5 Cong 0.9 654 900 #> #> $Incong #> cond perc time N #> 1 Incong 0.1 451 97 #> 2 Incong 0.3 521 291 #> 3 Incong 0.5 596 486 #> 4 Incong 0.7 680 681 #> 5 Incong 0.9 815 875 #> #> #> CAF: #> $Cong #> cond perc time acc N_A N_B #> 1 Cong 0.1 362 1 200 0 #> 2 Cong 0.3 426 1 200 0 #> 3 Cong 0.5 479 1 200 0 #> 4 Cong 0.7 549 1 200 0 #> 5 Cong 0.9 677 1 200 0 #> #> $Incong #> cond perc time acc N_A N_B #> 1 Incong 0.1 426 0.870 174 26 #> 2 Incong 0.3 518 0.995 199 1 #> 3 Incong 0.5 593 1.000 200 0 #> 4 Incong 0.7 676 1.000 200 0 #> 5 Incong 0.9 834 1.000 200 0 #> #> #> Parameter: #> Ter a c mu_t mu_f mu_RS2 mu_SS #> 1 0.2007522 0.2184259 0.2275249 0.08999085 0.1871032 0.4799693 0.7251469
or using plot()
plot(some_data)
Note that Sim_DDModel() initializes a simulation randomly by default, which means that the parameters within the model used for the simulation are uniformly drawn from the parameter domains specified in the model. If you want to run a simulation with specific parameter values, you must specify the parameters under the parameter argument! This argument requires a data.frame whose column names must be identical (in name and order!) to the parameters specified in the model!
custom_parameter <- data.frame(Ter= 0.2, a= 0.17, c=0.18 ,mu_t= 0.08, mu_f= 0.09, mu_RS2= 0.29, mu_SS= 0.5) custom_data <- Sim_DDModel(model = DSTP,trials = 1000L,parameter = custom_parameter) custom_data@PAR #> Ter a c mu_t mu_f mu_RS2 mu_SS #> 1 0.2 0.17 0.18 0.08 0.09 0.29 0.5 plot(custom_data)
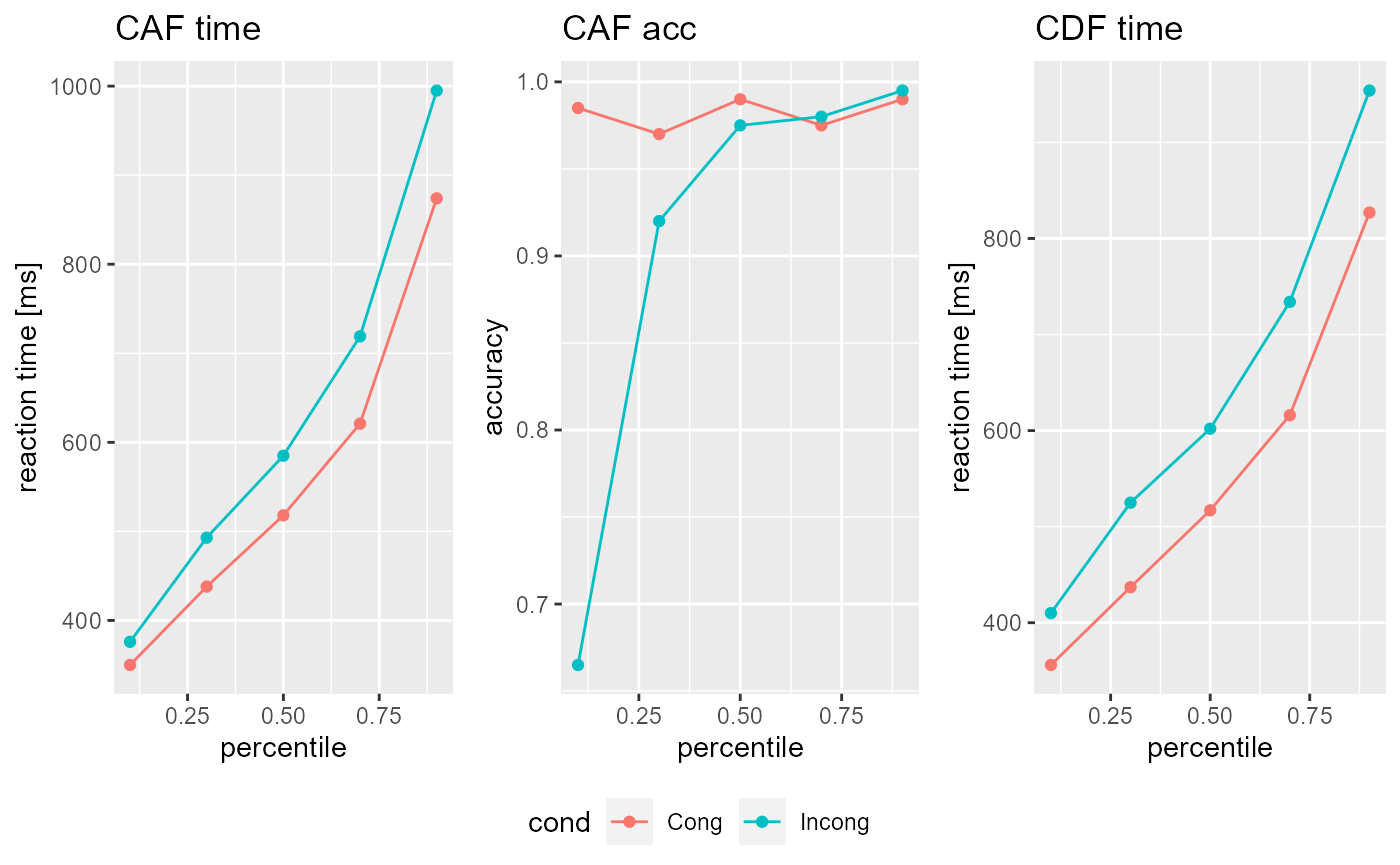
If you want to run several simulations with different parameters, just add rows to your data.frame! If you need multiple samples of these simulations, specify the simulations argument. Note, however, that Sim_DDModel() then returns a list of DDRep objects! If you do not specify the parameter argument, simulations=n>1 will result in n randomly drawn parameters as in the example above; but if you do specify it, simulations=n will result in n simulations for each parameter set!